Core Concepts
This article discusses protein sequencing and its techniques and applications in modern science.
Topics Covered in Other Articles
Introduction to Protein Sequencing
Protein sequencing is the process of determining the precise order of amino acids in a protein. Proteins, which are composed of long chains of amino acids, are fundamental to almost all biological processes. Proteins derive much of their function from their three dimensional structure, which is determined by the sequence of amino acids and how the protein folds. Therefore, understanding their sequence is essential, as even small changes in the sequence can dramatically alter their function. Protein sequences offer vital information in the fields of drug development, disease diagnosis, and evolutionary biology.
Traditional Protein Sequencing Methods
Traditional protein sequencing techniques laid the groundwork for modern day proteomics and provided critical insights into the primary structure of proteins. Two traditional protein sequencing methods include Edman degradation and mass spectrometry based sequencing.
1. Edman Degradation
One of the earliest and most widely used techniques is Edman degradation, developed by Pehr Edman in the 1950s. Edman degradation sequentially removes one amino acid at a time from the N-terminus of a protein. This allows the amino acid sequence to be determined step by step. The cleaved amino acid is then labeled and identified, while the remaining peptide chain undergoes further cycles of degradation until the entire chain is identified.
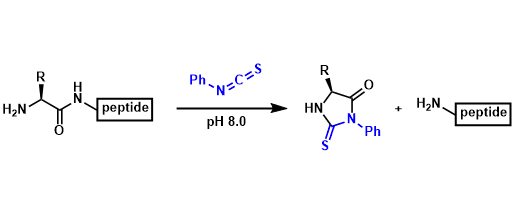
This method relies on the use of phenylisothiocyanate (PITC) to react with the free amino group of the N-terminal amino acid under mildly alkaline conditions. The reaction forms a phenylthiocarbamide (PTC) derivative. This is then cleaved off from the peptide chain as a thiazolinone derivative, leaving the remaining peptide intact. The thiazolinone is converted to a more stable phenylthiohydantoin (PTH) derivative, which can be identified using chromatographic techniques such as high performance liquid chromatography (HPLC). Although highly effective for shorter peptides (up to around 50 amino acids), Edman degradation has limitations with longer sequences, proteins with blocked N-termini or modified residues.
2. Mass Spectrometry Based Sequencing
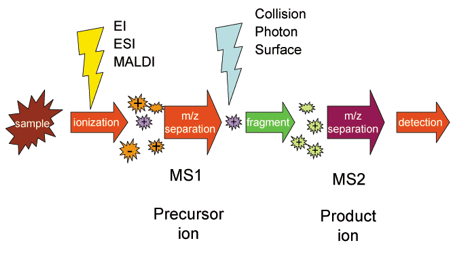
Mass spectrometry (MS)-based protein sequencing has revolutionized the field of proteomics due to its speed, sensitivity, and ability to analyze complex protein mixtures. In this method, proteins are first digested into smaller peptides, typically using enzymes like trypsin. Trypsin cleaves the protein at specific amino acid residues to produce predictable peptide fragments/ The resulting peptide fragments are then ionized, using techniques such as electrospray ionization (ESI) or matrix assisted laser desorption/ionization (MALDI). Once ionized, these peptides are introduced into the mass spectrometer, where their mass to charge ratios (m/z) are measured. Inside the instrument, peptides can also be fragmented further, producing characteristic fragmentation patterns that provide structural information about the peptides. By analyzing these patterns, lines are graphed onto the m/z spectral graph allowing for the amino acid sequence of the peptides to be determined. This information is then matched to databases to identify the original proteins. Overall, mass spectrometry works by separating ions based on their m/z values and recording their abundance, creating a spectrum that reveals both the composition and structure of the peptides as shown in the image below.

Tandem mass spectrometry allows for further fragmentation of these peptides, producing smaller fragment ions that can reveal amino acid sequence. Mass spectrometry based sequencing mainly focus on ionization processes, fragmentation pathways, and the detection of fragment ions. The combination of which provide a fingerprint of the peptide sequence. This approach is highly effective in identifying post-translational modifications MS sequencing can also handle a much broader range of protein sizes and complexities compared to Edman degradation.
Modern Techniques and Technologies
Because DNA encodes the instructions for protein synthesis, sequencing DNA can often provide the same information as sequencing proteins. High-throughput sequencing, often called next-generation sequencing (NGS), enables rapid and parallel sequencing of millions of DNA fragments. The process begins with DNA fragmentation, followed by adapter ligation and amplification through techniques like bridge amplification or emulsion PCR. Nucleotides are incorporated into the growing DNA strands, with chemical processes like pyrosequencing or reversible terminator chemistry detecting the sequence in real time. Advances in automation have significantly scaled up throughput, reducing cost and time while generating massive datasets. Sometimes, however, researchers may prefer to sequence proteins instead of DNA in order to study post translational modifications, which can’t be determined from DNA sequence alone. Additionally, the genome of the organism could be unknown or incomplete. In these cases, scientists revert back to the traditional methods of protein sequencing.

Another modern day protein sequencing technique is nanopore protein sequencing. This method analyzes single molecules by guiding them through a nanoscale pore (nano pore) embedded in the membrane. As the molecule passes through, an electric current flows across the membrane, creating measurable disruptions. For proteins, nanopore sequencing detects current changes based on the amino acid sequence, enabling real-time protein analysis. The nanopore, often made from biological proteins or synthetic materials, forms a highly sensitive channel. Each amino acid or modification causes a unique shift in current, which advanced computer algorithms decode. This method can sequence proteins in native or denatured states without extensive sample preparation, ideal for high-throughput sequencing.
Applications of Protein Sequencing
Protein sequencing carries vital real-world applications in medicine, biotechnology, and even agriculture. In the medical field, protein sequencing aids in the identification of genetic disorders and mutations as well as plays a key role in the development of vaccines. In pharmaceutical industries, protein sequencing aids in designing targeted drugs and therapies for diseases. Additionally, in agriculture, it improves crop resistance to pests and diseases. Protein sequencing also aids in producing biofuels and bioplastics in the field of synthetic biology. Further, it supports forensic science through protein-based identification methods. The potential for protein sequencing in science is seemingly limitless.
Limitations of Protein Sequencing
Protein sequencing techniques face limitations in accuracy and efficiency due to complexity and diversity of protein structures. There exists a reliance on high-quality samples which can hinder success, as degraded or impure samples often yield incomplete sequences. Current methods struggle to identify post-translational modifications, which play crucial roles in protein function. Many techniques also require large quantities of protein, making analysis challenging for low-abundance proteins. Additionally, protein sequencing can be time-consuming and costly, which limits its accessibility.
Mass spectrometry often has difficulties identifying proteins with similar molecular weights. Therefore, MS may not always detect small or subtle variations in amino acid composition. Edman Degredation on the other hand are unable to sequence proteins beyond a certain length. Fragmentation methods can introduce sequencing errors, especially for complex or highly similar proteins. Nanopore sequencing has issues with sensitivity to modifications, in which complex modifications can go undetected or misinterpreted. Overall, current sequencing methods lack the sensitivity for a comprehensive analysis of complex proteomes.
